B. Background
"I am the
relator in this action and make this declaration in opposition to defendants’
motions to dismiss the First Amended Complaint on jurisdictional grounds
pursuant to Rule 12(b)(1) of the Federal Rules of Civil Procedure. I have personal knowledge of the facts
set forth herein, and am willing and able to testify to them under oath." ... (First
Decl. par. 1; see footnote 2 for reference abbreviations).
"To assist
the Court in making the necessary findings regarding the absence of any public
disclosure of the relevant allegations or transactions, and particularly to
assist the Court in understanding the nature and extent of my role in
investigating and developing the allegations in the Original and First Amended
Complaints, I believe it is essential for me to provide the Court with some
background information about myself, and about the scientific research which
was the subject of the grant applications in connection with which the
defendants made the false claims at issue in this action." (First Decl. par. 3.)
"My
educational background is as follows. I received an A.B. degree in Biochemistry
from Harvard University in 1982. Thereafter, I completed a joint M.D./Ph.D.
program at defendant Cornell University Medical College (“Cornell”) and the
Sloan-Kettering Institute. I received my Ph.D. degree in Molecular Biology from
Cornell in 1990, and my M.D. degree from Cornell in 1991." (First Decl. par. 4.)
"From
approximately 1987 through 1990, while pursuing my Ph.D. degree, I worked as a
graduate student in the laboratory of defendant William K. Holloman, Ph.D.
(“Holloman”) at defendant Cornell.
At the time, most of the research conducted in the Holloman Laboratory
was focused on a gene
known as REC1 and, to a lesser
extent, on a gene known as REC2. During
the period that I worked in the Holloman laboratory, I, along with others, was able
to successfully isolate the genes for REC1
and REC2. My work as a molecular
biologist in the laboratory also included characterizing and obtaining sequence
from those genes. In
addition, I had extensive interaction with others in the laboratory who were
attempting to reproduce and extend the Rec1 work of Kmiec and Holloman,
including by my attending laboratory meetings in which these issues were
discussed. Therefore, my status as an “insider” of the Holloman laboratory gave
me extensive knowledge and access to issues regarding Rec1/REC1 and Rec2/REC2
research." (First Decl. par.
5.)
"From 1991
through 1996, I continued to pursue research related to that performed in the
Holloman lab while a
Postdoctoral Fellow in the Columbia University Department of Genetics and
Development." (First Decl. par. 6.)
"The work I
performed on the REC2 gene during the
period of my postdoctoral fellowship was related to demonstrating that the rec2-1 gene was a functional null. Based
on observations made during that work, I also obtained evidence concerning the
mutator status of rec2 mutants.
Finally, I sequenced upstream regions of the REC2 and rec2-1 genes.
(These sequences are directly relevant to this case and which establish my
position as an original source for this vital information.)" (First Decl. par. 7.)
"From 2001
to [October 2007], I [was]
employed by the St. Luke’s-Roosevelt Institute for Health Sciences at Columbia
University as Director of the Cognitive Neuroscience Laboratory. From 2000
through 2002, I assisted science journalist Gary Taubes with an article
on various phases of concern in the scientific community over the work of the
Defendants. The phases of such concern are summarized in the following
paragraphs." (First Decl. par. 7
[sic - the second of two paragraphs numbered "7"].)
"From the
early 1980’s through the mid-1990’s. Holloman and his graduate student, Kmiec,
claimed to have been the first to purify and study a protein,
Rec1, which they identified as coming from a gene known as REC1. This protein had special properties common to a class known as DNA recombinases. However, only Kmiec could
produce the purified Rec1 activity, despite prodigious efforts by many others
in the Holloman laboratory and outside it. This situation was of such
longstanding concern in the scientific community that it became public enough
to be described in a Science magazine article published in 2002. I term this
the “Rec1 Phase” of the controversy
and disputes noted by the Defendants." (First Decl. par. 8.)
"Upon
leaving Holloman’s laboratory to perform a postdoctoral fellowship in the
laboratory of Dr. Abraham Worcel, Kmiec seemingly replicated his remarkable
Rec1 feats by producing data ostensibly showing that he had purified another
important eukaryotic enzymatic activity known as a gyrase. The activity Kmiec
purified had already been known as TFIIIA. After sustained objections from a
second laboratory challenging Kmiec’s TFIIIA results, Dr. Worcel had numerous
members of his laboratory attempt to replicate Kmiec’s results. They could not
do so. In stark contrast to Holloman’s relentless and self-serving support of
his irreproducible Rec1 work with Kmiec, Worcel issued a very public retraction
of the work which Kmiec had done on TFIIIA in his laboratory).
I term this the “TFIIIA Phase” of the
controversy and disputes noted by the Defendants." (First Decl., par. 9.)
"Phases I
(Rec1) and II (TFIIIA) were based upon purification of proteins (Rec1 and
TFIIIA) and study of their biochemical activities. The outcome of those two
phases ended negatively enough that Holloman told me when I was a graduate
student in his laboratory that he was “under a cloud”, of suspicion by other
scientists. The reason was quite apparent. If the TFIIIA work was a near certain
fraud by Kmiec which had to be retracted by his employer, and no one had
replicated the Kmiec Rec1 work in nearly a decade of attempts, then it would be
natural to assume that this work was fraudulent also." (First Decl., par. 10.)
"Holloman
also apparently understood that a defense to views against the veracity of his
prior Rec1 publications would be the determination of its gene sequence. As
noted above, I and others were able to successfully isolate the gene for the
Rec1 protein, i.e. REC1. Holloman’s
hopes for sequence salvation were dashed, however, when it became apparent that
the REC1 DNA sequence would not
specify the protein elements common to known recombinases, such as Rec1 was
purported to be. However, I and other molecular biologists working for Holloman
had also cloned and sequenced a second recombination-related gene known as REC2. “As fortune would have it”,
Holloman noted in one of the grants at issue in this case, the REC2 gene did have a sequence consistent
with a recombinase. However, among other problems, Holloman and Kmiec had in
two earlier publications produced data linking the Rec1 protein activity as
emanating from the REC1 gene and
explicitly not from REC2. Therefore,
Holloman was faced with the need to transform the disputed Rec1 protein
activity claimed by himself and Kmiec into the Rec2 protein, which undoubtedly
did exist and would be predicted to have recombinase activities. It is three
specific [alleged] fraudulent actions that the Defendants took to effect this
transformation of irreproducible Rec1 into the predictable Rec2 recombinase
that are the focus of this case. I term this the “Rec1-is-Rec2 Phase” of the controversy and disputes noted by the
Defendants." (First Decl., par.
11.)
"Beginning
in a 1996 work, and continuing to the present, the Defendants (primarily Kmiec)
have published research on a technique related to the earlier phases (the
recombination and repair of DNA), but with more direct therapeutic potential.
This technique, termed by them “chimeraplasty”, has been the subject of
substantial public controversy, including as the primary focus of the
aforementioned Science magazine article. However, we do not make allegations in
this action regarding this [fourth] phase of concern regarding the work of the
Defendants, other than to note that it is our understanding that Defendant
Thomas Jefferson University was asked to investigate Kmiec’s chimeraplasty work
while he was employed there, but turned this request down.
Nevertheless, Dr. Kmiec was a known issue to the university. (Repeated red
flags and warnings to Defendant Cornell about Holloman will be detailed at
trial."
(First Decl., par. 12.)
Back to TOC.
C. Basis for the Allegations
"...
on or about November 24, 1994, in a conversation with a colleague, Hamish
Young, a professor at Columbia University who had worked with me [while he had
been] on sabbatical in the Holloman lab ... [Dr. Young] informed me that a “Rec1 is
Rec2” paper had been published by defendant Holloman. See
Bauchwitz Dep. 240:1-245:23.
That paper turned out to be the 1994 Paper. Dr. Young also told me that defendant
Holloman had been taking “flak”, so that the “Rec1 is Rec2” paper represented a
“vindication.” My notes show that I
had considered such a claim quite unexpected. Id."
(Sec. Decl., par. 13.)
"Upon
hearing [] from Dr. Young that the “Rec1 is Rec2” findings had been published, I
obtained a copy of the 1994 Paper in or about late November or early December
1994. I immediately noticed that Brian
Rubin – my successor in the Holloman lab -- was not listed as an author,
which I found to be both unusual and disturbing, given my own prior negative
experiences in scientific publishing with defendant Holloman (as summarized in
Table 2 of Plaintiff’s Statement of Additional Facts, and in Answer to Interrogatory
No. 18 of Plaintiff’s Answers to Jefferson Interrogatories). As a result, I contacted Rubin and had
two telephone conversations with him about the 2004 Paper – the first on
December 27, 1994, the second on February 13, 1995. I tape-recorded both conversations, and
later (in February 2004) transcribed and annotated these two recordings
myself." (Sec. Decl., par. 14.)
False Claim 1: Rec1 protein sequence fabrication/falsification
"In
our conversation on December 27, 1994, Rubin informed me that, despite working
on the issue for a period of time, he and another post-doctoral fellow in the
Holloman laboratory, Naoto Arai, had been unable to reproduce the results
indicated in the 1994 Paper, and, given Kmiec’s prior history, did not agree
with its conclusions and did not want their names listed on the 1994
Paper. As Rubin stated in our
December 27, 1994 conversation:
Rubin: “So he [postdoctoral fellow Naoto Arai] came, and he tried to
figure out if the Rec2 protein was a strand exchange protein. He worked on it
for, well, almost two years. I'd say about two years. And he got the same results I did. Basically, we could never show this
did anything. And we purified numerous helicases from Ustilago and E. coli, and
basically we were looking for a DNA dependent ATPase. We tried to follow,
that was sort of our base assay, was DNA dependent ATPase. We also did
extremis. We tried strand exchange on
our purified preps, but we never got it to do anything. …
But, so in the meantime, what
Bill did was he sent my overpurification, Ok, so he sent my overexpressing
strain to Eric's [Defendant Kmiec’s] lab. Because, in my heart, I kind of believe what happened was with him was,
he just really wanted this result. To be a strand exchange protein. And I think what he did was he
figured, well, these guys in my lab are not getting this. So I am going to send
it to Eric because Eric was starting to get back into Ustilago research. So he
sent him our overexpression strain. And
then Eric came up with this incredible result that, he purified Rec1 protein
from Ustilago and sent it to Harvard for sequencing, and several of the
peptides were identical to Rec2 sequence. I was very skeptical about all this.”
See Transcription of
Recording of December 27, 1994 Telephone Conversation with Brian Rubin,
attached as Exhibit N to Defendants’ Joint Statement of Undisputed Facts [at
RPG 00009-00010]." (Sec. Decl.,
par. 15.)
"Rubin
stated, he was “very skeptical about all this”. (emphasis added). I too was skeptical
about the results reported in the 1994 Paper, but note that elevated skepticism
about the results reported in a published research paper is not the
equivalent of knowledge that a fraud had occurred, or even evidence of such."
(Sec.
Decl., par. 16.)
"As
I testified at my deposition (see Bauchwitz
Dep. 131:21-138:15), if the situation had [] only been that Rubin and Arai couldn’t get the same result that
Kmiec purported to have gotten, I would NOT have even called ORI in 1995, as I
would not have expected this to be sufficient basis for them to [] investigate.
It was only in the context of their prior pattern of dishonest behavior,
which preceded and was separate and distinct in nature from the current
information I was hearing for the first time, that I felt an investigation
by ORI was warranted in 1995. Clearly, this same consideration of a prior
pattern of questionable performance had influenced Rubin, as he noted in the
same conversation:
“This totally disagrees with my
conclusions and, knowing the nature of Eric having had several problems in
science, of reproducibility, I said "Just take my name off the
paper."
See Transcription of Recording of December 27, 1994 Telephone Conversation
with Brian Rubin, attached as Exhibit N to Defendants’ Joint Statement of
Undisputed Facts." (Sec. Decl.,
par. 17.)
"As
Rubin stated, “knowing the nature of Eric [defendant Kmiec] having had several
problems in science [irreproducibility of Rec1 and TFIIIA protein activities]”.
The pattern of prior behavior by the Defendants was integrally involved in
interpreting the current unlikely scenarios that I was hearing." (Sec.
Decl., par. 18.)
"As
a result, the information that I received from Rubin in their conversation of
12/27/94 did not constitute notice of a likelihood of fraud." (Sec.
Decl., par. 19.)
"Upon
being contacted by me in 1995, the ORI could have done an investigation and
perhaps have developed such information. To start, it would have been able to
interview Rubin and me, as well as transcribe my recordings if required.
However, as it turned out, ORI apparently did not investigate." (Sec.
Decl., par. 20.)
"As
the transcript of my December 27, 1994 conversation with Rubin makes clear,
Rubin did not say anything to me to suggest that any of the defendants had
sought or obtained grant funding based on the 1994 Paper. There was no basis for me to be watching
for future grant applications containing “false” statements based on the 1994
Paper, since I did not have adequate knowledge that they were false." (Sec.
Decl., par. 21.)
What
I eventually did, after learning of the later false claim published by
defendant Holloman in the 2001 Paper, and deciding to investigate further, was
to reconsider and analyze the circumstances of the Rec1 protein sequence in
more detail. It occurred to me that for the Rec1 protein sequence to have
been as the Defendants claimed (identical to that expected for Rec2), data
might have been available at a laboratory external to theirs which had
performed part of the work claimed. In other words, as I explained in detail in
my July 26, 2004 FCA Disclosure to the ORI (Exhibit U to Defendants’ Joint Statement of Undisputed Facts), and in
the First Bauchwitz Declaration, it might be the case that purified Rec2
protein made from bacteria and submitted as if Rec1 protein purified from fungi
would show evidence of bacterial rather than fungal protein background.
(Sec.
Decl., par. 22; emphasis added.)
"To pursue this possibility,
on April 30, 2003, I contacted the Harvard Microchemistry laboratory, the outside
laboratory defendants claimed to have employed. See
Bauchwitz Dep. 296:21-298:23. Amazingly,
I never even got to the point at which data could be identified for subsequent
analysis (bacterial vs. fungal origin). Instead, I received information that led
me to conclude that there was a very high likelihood that the Defendants had
never sent anything at all to that laboratory in the relevant time frame.
Consequently, at that moment in 2003,
a suspicion about whether Rec1 could
have been Rec2, which had been reported to authorities in a timely manner in
1995, had upon my investigation led to
evidence of a potential fraud - a lie." (Sec.
Decl., par. 23.)
Back to TOC.
False Claim 2: falsification of rec2-1 mutant DNA sequence
"As Rubin and I discussed in our February 13, 1995 telephone
conversation:
Bauchwitz: “Another question I had out
of curiosity, this is probably very simple but, you show the sequence
and the map of the deletions. Where is the methionine, the ATG,
where would that be that you could start the truncated message? Because the the
Kmiec paper they give the whole story about how this rec2-1 smaller message
produces a smaller truncated protein.”
Rubin: “Oh, OK. So its
methionine is gone. What they would postulate is that you
picked up a methionine that is downstream of the deletion. So it just basically
starts at the first methionine.”
Bauchwitz: “ … there aren't any
methionines until after the recA and Walker A homology sites. In fact it is two
amino acids after, there is not even AT [or G except] immediately after the []
site.”
Rubin: That's interesting. I
never really thought about that but,”
Bauchwitz: “Oh so there wasn't
actually a methionine in mind.”
Rubin: “Oh no, no, they wouldn't,
they hand
waved. If you brought that, what you just told me up, I think what they
would probably say is "Oh, it must read an upstream methionine then, that
is basically to the left of the deletion."
See Transcription of
Recording of February 13, 1995 Telephone Conversation with Brian Rubin,
attached as Exhibit P to Defendants’ Joint Statement of Undisputed Facts [at
RPG 00001-00002]." (Sec. Decl.,
par. 25.)
"This
key interchange clearly indicates that Rubin had not even considered the relevance
of the rec2-1 start site as being in a position in which it would have
eliminated activity from the protein which was claimed by Kmiec and Holloman,
in seeming contradiction to earlier data, to be active (in vitro). Rubin’s response to me was that “hand waving” would have
been the likely response of Holloman and Kmiec to the questions I was
raising." (Sec. Decl., par. 26.)
"Rubin’s
characterization of the likely response of Holloman and Kmiec to my questions
as “hand waving” did not suggest to me any likelihood of fraud. Clearly, my conversations with Rubin did
not suggest or place me on notice of the second alleged fraud (falsification of
rec2-1 sequence)." (Sec. Decl., par. 27.)
"What
is much more important in the conversation with respect to this second fraud is
when I asked Rubin where to find his rec2-1 sequence (and the comparable region in
REC2), as noted in this exchange:
Rubin: You know
what is funny is, that somehow the, I actually did notate that the
rec2-1 mutant, that on this figure originally, Figure 1, and uh, in the legend,
5 the description, I am looking at my thesis right now, and it was the same
figure, and it is in there.
Bauchwitz: Oh, it is? Rubin:
Bill
must have taken it out for some reason.” (Sec. Decl., par. 28.)
"So
it seemed to Rubin an oddity that Holloman had removed the sequence of interest
from his own 1994 publication with Rubin (a distinct publication from Kmiec et.
al., 1994, from which Rubin had removed his name). In 2005, ORI would note to
the Department of Justice that Holloman many times alluded to having published
the relevant sequence but did not actually do so (See ORI Review of Complaint at 5, Exhibit TTT to Defendants’ Joint Statement of Undisputed Facts).
Indeed, even the comparable region of REC2 (wild-type gene) was not given in
Rubin’s thesis. These numerous omissions by defendant Holloman made me suspicious
enough that I endeavored to produce the sequence myself, which I
published in the federal Genbank database in 1999. Again, suspicion led to
investigation. Investigation led to information that put me in a position to
perceive a lie (fraud) when it was eventually published in 2001." (Sec.
Decl., par. 29.)
"Why
was Holloman seemingly hiding the relevant rec2-1 sequence? Neither Rubin
nor I knew in 1995 that in 2001 Holloman would publish an explicit
falsified statement about that sequence as part of a cover-up of the
Rec1 is Rec2 situation. See Bauchwitz First Declaration, ¶ 26."
(Sec.
Decl., par. 30.)
"In
fact, if I had not produced and by 1999 published my own relevant rec2-1 sequence, in
2002 I still would only have had suspicions and concerns about what Defendant
Holloman had written in 2001, since I would not have had access to any
sequence from Rubin (1994 Paper or 1995 sequence) or Holloman (Genbank database
– no actual entry; e.g. see Exhibit
7 to Plaintiff’s Statement of Additional Facts [Doc. 90]). Therefore, it was
my own investigation of the rec2-1 sequence which gave me the specialized
knowledge that enabled me to detect and understand the patent lie published in the
2001 Paper. It was at that moment that this qui tam case began. See Bauchwitz First Declaration, ¶¶
26-28." (Sec. Decl., par. 31.)
"The
only question with respect to alleged frauds 1 and 2 would seem to be whether
I had any duty in 1995 - long before I even knew what a qui tam action was, or had even
considered whether defendants might seek future
federal grant funding based on the False Claims 1 and 2 – to act as a
perpetual investigator and monitor of future grant applications by the
defendants, based solely on my skepticism about the results
reported in the 1994 Paper. I
respectfully submit that that this cannot be the case. To the contrary, I believe that in 1995 I
went above and beyond the call of duty by notifying ORI of continuing
suspicious circumstances that warranted investigation, and subsequently
developing my own rec2-1 sequence, when nothing by Rubin could be found." (Sec.
Decl., par. 32.)
"Once
I developed solid evidence of lies -- of frauds -- as I understood them, in the
spring of 2002, I took very timely action to obtain
relevant grants by the end of 2002. My continued investigation led to
additional findings of fraud in 2003 and 2004, resulting in the filing of my
original Complaint on June 30, 2004, less than two and a half years after
I first saw what I knew to be a published false statement by one of the
defendants (in the 2001 Paper)." (Sec. Decl., par. 33.)
"In
1995, I more than discharged any duty on my part to act by alerting ORI to suspicious
circumstances. I did not give them specific allegations of fraud
because
I myself did not have them until 2002, as detailed above. ORI has
clearly stated the same, i.e. that I never gave them specific allegations of
fraud until our meeting in July 2004 (see
ORI Time Line, Exhibit 10 to Exhibits to Plaintiff’s Statement of Disputed
Facts and Plaintiff’s Statement of Additional Facts [Docs. 89 and 90])." (Sec.
Decl., par. 34.)
Back
to TOC.
False Claim 3: falsification of Rec2 protein activity
"In
our December 27, 1994 telephone conversation, Rubin also made the following
comments:
Rubin: “I think of him as just a
guy that develops stories in his office and then comes into the lab and says
produce the data that fit my stories. And in fact in that paper, there are figures
straight from my thesis that have totally nothing to do, really, with what was
published.”
Bauchwitz: “What do you mean by
that?”
Rubin: “Well like the protein, he
[Defendant Holloman] used my purification gel and Western showing the anti-Rec2
antisera binds this protein.”
Bauchwitz: “Yeah.”
Rubin: “And then he claims that this protein
is active. And he would rationalize it by saying ‘Well, it's
just a better looking gel than Eric's [Defendant Kmiec]’ Of course, we are
using the same strain, but that prep wasn't active.
Bauchwitz: “But the gel is of
yours, then it really isn't of the same prep that he used to show
activity.”
Rubin: “Exactly.”
Bauchwitz: “He was accused of that,
I
am not sure if that is the right word, but Toyoko
said that he did that repeatedly with Rec1. There would be some crude fraction
of unknown meaning that had some activity that he was ascribing to some purified
band.”
Rubin: “Exactly.”
Bauchwitz: “A decade ago.”
Rubin: “This is more of the same.”
See Transcription of
Recording of December 27, 1994 Telephone Conversation with Brian Rubin,
attached as Exhibit N to Defendants’ Joint Statement of Undisputed Facts [at
RPG 00011-00012]." (Sec. Decl.,
par. 35.)
"The
third fraud claim alleges falsification of a data image in the 1994
publication. The key issue as to whether I was put on notice of this claim from
my December 1994 telephone conversation with Rubin is what I derived (or a
reasonable person in my position would have derived) from the
conversation. My language in the
call indicates that Rubin’s passing comments about his gels did not trigger any
immediate suspicion of fraud. Neither Rubin nor I mentioned the word “fraud”.
Instead, both Rubin and I concluded that the behavior was “more of the same”, i.e.
consistent with behavior which had generally been associated with Defendant
Holloman by those who had worked for him. We then returned to the topic of the
call, namely how Rec1 could be Rec2 and why Rubin (and Arai) had removed
themselves as authors from this claim. While I do not recall mentioning any of
this information about the misuse of Rubin’s gels in my call to ORI in early
1995, I believe that I more than discharged any duty to generally bring
information of the suspicious circumstances surrounding the 1994 Paper to the
attention of authorities (ORI)." (Sec. Decl., par. 36.)
Back to TOC.
D. Timeline and Handling of the Case by the Office of Research
Integrity
During
the summer of 2000, a journalist, Gary Taubes, contacted me regarding an
investigative article he was writing about the (future) defendants, for which
Taubes sought assistance. Upon completing his work (Taubes2002Chimeraplasty Exhibit, in the Case Documents folder on
the CD), the journalist suggested to the federal government’s Office of
Research Integrity (ORI) that they contact me to consider a misconduct
investigation.
I was leery of
working with the ORI based on my prior experiences in reporting concerns about
possible misconduct to them and their predecessor, OSI. Those contacts were
summarized in tables in the case document, Additional
Facts (also in the Case Documents CD folder):
Table 2. STATEMENTS TO OSI 1990
|
Statement
|
|
Type
|
Research Topic
|
|
|
1
|
Harm
to students by Defendant Holloman, apparently because they could not
adequately reproduce potentially irreproducible work by an earlier graduate
student (Defendant Kmiec) who had his subsequent postdoctoral work retracted
|
Serious
concern
|
Rec1
protein purification and activity
(“Rec1”)
|
|
|
2
|
Misconduct
in research publishing by Defendant Holloman
|
Allegation
|
REC1
cloning, REC2 cloning
|
|
|
3
|
Were
negative data omitted in claims made in new NIH grants?
|
Suspicion
|
rec2-1
homologous integration issue
|
|
Table 3. STATEMENTS TO ORI 1995
|
Statement
|
|
Type
|
Research Topic
|
|
|
1
|
What
became of the concerns and suggestion for investigation made to OSI in 1990?
|
Follow
up
|
1990 1-3 (see Table 2)
|
|
|
2
|
Defendants
have now published a paper under highly suspicious circumstances, based on
results purportedly obtained by Defendant Kmiec but which two members of
Defendant Holloman lab were unable to obtain. (Reminiscent of the Rec1
situation reported to OSI in 1990.) Prior scientific findings are not
consistent with the published claims and suggest the new claims are highly
unlikely to be true.
|
Suspicion
|
Regarding
research publication: Kmiec et. al., 1994 (MCB)
(“Rec1
is Rec2”)
|
|
"Regarding
the events which specifically led to this legal action, I provide the following
timeline: On February 9, 2002, science journalist Taubes brought
to my attention a new article published by Defendant Holloman (Kojic et. al.,
MCB, 2001, hereinafter, Kojic et. al. 2001) which pertained to a mutant DNA
sequence of interest in this case (“rec2-1”).
After eventually obtaining and reading Kojic et. al., 2001, I saw what I knew
to be a clearly false statement made by Defendant Holloman regarding the mutant
sequence:
“Inspection of
the rec2-1 DNA sequence on both sides
of the deletion indicated that a novel ORF could be generated through
conjunction of the flanking sequences. This ORF would be predicted to encode a
613 amino acid Rec2 protein variant with a novel 19-residue leader sequence
derived from upstream of the wild-type Rec2 protein.”
I also realized that this false
statement was meant to provide cover for the earlier incredible claims made in
Kmiec et. al., 1994, (namely that the Rec1 protein activity had actually
emanated from the REC2 gene).
Specifically, the motivation for this false statement was to explain explicitly
how the Rec1 protein Defendant Holloman had originally published with Defendant
Kmiec could have been derived from a different gene (the REC2 gene), despite genetic and other evidence to the
contrary." (First Decl. par.
26.)
"Furthermore,
according to information elicited by me from Rubin, Holloman knew of the true
state of the mutant gene in question prior to his false representations in
2001. ... " (First Decl. par.
38.)
"... On
April 30, 2003, I called the Harvard University Microchemistry Facility. I
spoke with intake specialist Liam McCallum. McCallum told me that the last
record for Kmiec was a user id on December 31, 1992. With respect to the other
two authors with Defendant Kmiec, there were no records for Allyson Cole, and
the first record for Defendant Holloman was not until 1995 (after the date of
acceptance for publication of Kmiec et. al. MCB 1994 on July 28, 1994).
McCallum noted that the Kmiec record had "no activity on it". He said that it remained their practice to
give anyone who contacts them a user id. Contemporaneous records from other
researchers did indicate activity. He noted that it was possible that further
records, if any, (such as invoices sent for services rendered) existed at an
"off-site storage area”. I felt that this information was
obviously consistent with the amino acid data having been fabricated.
(First Decl. par. 27.)
As it turned out
during discovery, Harvard had indeed retained complete paper records in its
archive facility. Their records indicated that protein microsequencing had been performed at Harvard for Kmiec
during the period in question. The records also indicated that the reason the
intake specialist did not see relevant entries was that the electronic database
had been changed shortly after that time.
Nevertheless,
and most importantly, the documents released by Harvard also clearly indicated
that the Harvard Microchemistry
laboratory had not produced the data claimed by Kmiec and Holloman in their
paper KCH1994, and in associated federal grants. [See New Evidence, Section
E., below.]
The Court has
argued that I had a duty to call Harvard's Microchemistry Laboratory within a
specified time of being put on notice of fraud. As noted above, it is arguable
as to what conclusions about fraud were reasonable at the time of the reading
of the 1994 publication and speaking with Rubin. However, it is also important to note that when I
ultimately did contact Harvard's Microchemistry Laboratory, its director did not release any
records nor even confirm that his facility had produced the findings claimed.
Therefore, contrary to the Court's
implication that such information would be readily available for those who
contacted the third party, in reality the use of subpoenas was
ultimately vital to resolving the nature of what data had or had not
been obtained.
With respect to
the third allegation, "After many months of attempting to have commercial
firms provide the transcriptions, I myself completed them on January 27,
2004." (Sec. Decl., par.
14.)
Ultimately,
ORI agreed to proceed on the basis of a qui
tam suit. The case United States ex
rel Bauchwitz v. Holloman et. al., No. 04-2892 (E.D. Pa.) was filed under
seal on June 30, 2004 in federal district court in the Eastern District
of Pennsylvania (the location recommended by the ORI). The ORI was to produce a
report for the Department of Justice on the science involved.
Back
to TOC.
ORI Statements and Responses
ORI
produced a report for use by the Department of Justice (DOJ), which was
received by me via DOJ on January 12, 2005. In that report, the first of
what collectively have come to be called the “ORI documents”, ORI concluded that each of the Plaintiff’s
allegations had merit.
However,
ORI also noted what they believed could be two primary issues in pursuing the
case: 1) ORI thought there might be no additional evidence available beyond that
which the Plaintiff had brought to the government, and 2) ORI claimed that it
might not be possible to prove intent for one of the false claims alleged. (The
complete ORI documents are available at court docket Doc. 90 attachment 9 and
Doc. 90 attachment 10 of 4/16/08 and on the enclosed CD).
The
following are excerpts from the conclusion of ORI's first memorandum of November
23, 2004 [with emphasis added]:
i. "Each [of the Relator's claims]
has some merit, but all lack definitive proof of being deliberate
falsifications, and ORI does not believe that evidence is available to provide
such proof."
ii. “Dr. Bauchwitz’ complaint
identifies three false claims, as identified above. ORI notes that these false claims
deal with only a very small portion of the much larger scope of possible misconduct
issues that have been linked to Drs. Kmiec and Holloman (see footnote
8). The reason for this is that Dr. Bauchwitz has limited his claims to issues
that he has direct knowledge of. He has made a solid case that the ‘story’ on Ustilago maydis recombination genes,
their associated proteins and their enzymatic properties has shifted
dramatically over the past 20 years. Many scientists working in this area
appear to have believed that erroneous
claims have been consistently published by Drs. Holloman and Kmiec.”
iii. "Even if it could be
shown that some of the grant applications unequivocally contain the false
statements described in the complaint, ORI believes that the evidence is
inadequate and generally unobtainable to prove that the questioned statements
are intentionally false".
With
respect to ORI's purported concern about not finding additional evidence, my
attorney responded that ORI had made no attempt to obtain such
evidence. There was then correspondence back and forth as to the
general lack of investigation and what should have been done to investigate.
Excerpts of communications and reviews related to the ORI's handling of the
case and general failure to investigate, are provided in the endnotes (i).
Ultimately,
contrary to the ORI's stated concerns, additional, important evidence was obtained during the limited
discovery that was performed in the case. That evidence and its assessment are
presented in the following sections.
Back
to TOC.
E. New Information Obtained During General
Discovery
I. Amino Acid
Sequence Data Fabrication/Falsification - First Allegation
Holloman
has claimed repeatedly that he has identified the Rec1 strand exchange activity
[KSSEP]
as being produced by the Rec2 protein. The claim is based on peptide sequencing
he and Kmiec published in Kmiec, Cole, and Holloman, 1994
("KCH1994"):

Selected “Rec1 is Rec2” Grant Claims making similar
claims (as anonymized for expert reviewers):
“From one, peptides were
isolated after digestion with trypsin and amino acid sequencing was performed
on five isolated peptides. From the second, N-terminal sequencing was carried
out. All the amino acid sequences obtained were found to be contained within the
R2 open reading frame (K et. al., 1994,
see Appendix)”.
GM042482-08 1996
– 2001 $3,
487, 424
“We obtained amino acid
sequence of peptides derived from the 70 kDa protein purified from fungus
XX. As fortune would have it the sequence information obtained corresponded
precisely to R2, a gene which had also been cloned and was under study in the
laboratory (R and H, 1990; B et. al., 1994; K et. al. MCB 1994; Appendix)”
GM53732-01 1996
– 1999 $1,127,533
“However, despite the name and the absence of detectable
activity in extracts from the r1 mutant, the R1 protein is encoded by the r2
gene (25).”
AR44092-01 1996
– 2000 $432,
657
The
amino acid sequences claimed in Kmiec, Cole, and Holloman, 1994 are shown in a
figure from that publication:

As
reviewed in the Basis for the Allegations section, above, I considered the
“Rec1 is Rec2” claim highly suspect for several reasons. Among the most
important reasons was that “Rec1” [KSSEP] activity was found in rec2-1 mutant
cells, but not rec1 mutant cells.
Yet rec2-1 mutants appeared to be functional nulls when I had worked
with Holloman. My sequencing of the rec2-1
allele after I had left Holloman’s lab had strongly indicated that any protein
fragment produced by it would not have strand exchange activity. (See also
Section E.II., below.)
Therefore,
records of the peptide sequencing of Rec1 strand exchange protein claimed in
KCH1994 were sought during general discovery in the qui tam science fraud case.
The
Harvard Microchemistry Laboratory had been cited for peptide sequencing in
KCH1994, as had Thomas Jefferson’s Cancer Center. Only Harvard’s Microchemistry
facility produced relevant research records. Kmiec had been employed by Thomas
Jefferson University.

In
a letter from Holloman to William S. Lane, Director of the Harvard
Microchemistry Facility on July 10, 2007, Holloman identified the date on which he and Kmiec claim that Harvard
performed the microsequencing
presented in KCH1994.

Holloman
also provided to Lane an HPLC profile of tryptic peptides,
labeled with “trp” designations, which Holloman claimed his records indicated
that Harvard had produced for him and Kmiec on September 10, 1993.
The
profile of the peptides sent to Lane by Holloman is shown below. It is
important to note that someone
had labeled five
of the peaks "trp1" through "trp5". Those designations
correspond to the peptides listed in Holloman's letter to Lane, and to what
Holloman and Kmiec claimed in their 1994 paper that Harvard had produced for
them.
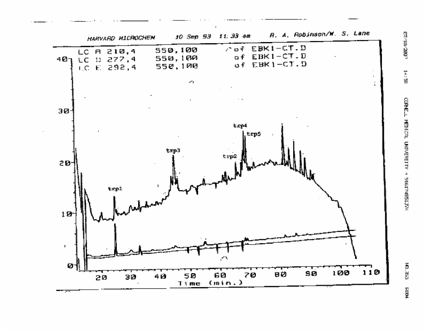
However,
documents obtained from Harvard University during discovery showed that only
three peptides had been sequenced, not five as claimed by Holloman:

The
actual data produced by the Harvard Microchemistry Laboratory is shown below.
It does not show the five printed "trp" labels Holloman had on his
version of the data, but rather has check marks over three of the peaks
(numbers 34, 61, and 73).

EBK1-CT34
amino acid data from Harvard Microchemistry

EBK1-CT61
amino acid data from Harvard Microchemistry

EBK1-CT73
- no amino acid data observed - from Harvard Microchemistry
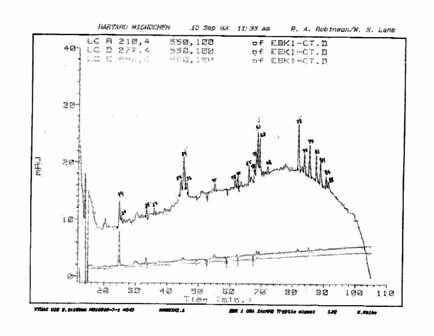
In
comparing the profile sent by Holloman to Lane with the profile Harvard claims
it actually produced, it can be seen that:
1) peaks 34 and 61 correspond to those labeled by
Holloman and Kmiec as “trp3” and “trp4”,
2) peak 73
was not listed by Holloman as having been sequenced, and,
3) Harvard does not note having sequenced peaks labeled
“trp1”, “trp2”, or “trp5” by Holloman.
Of
even greater importance is that Harvard also produced copies of the actual
sequence data that they had provided to Holloman and Kmiec in 1993.
They gave this information to Holloman in 2007 in response to his inquires
(e,g, the letter noted above), and to the court handling the False Claims Act
action in 2010.
Copies
of the sequence data for the three peaks is provided in the file, "Harvard Amino Acid Sequences" (on
the CD). Also shown in those documents is a data form indicating that Harvard
had tried to directly sequence the (presumably Rec1/KSSEP) protein as received
from Kmiec. However, Harvard obtained no sequence, which is what led to their
digesting and purifying fragments from the protein Kmiec had sent to them. (All
of the samples have an "EK" or "EBK" designation.) Thus, tryptic peptides
from sample EK1 were subsequently HPLC purified to produce peaks EBK1-34, -61,
and -73.
The following list summarizes the amino acid sequences
claimed by Holloman in his letter to Harvard, those that were actually produced
by Harvard upon subpoena, and those published in Figure 1 of Kmiec, Cole,
Holloman 1994:
trp1 DAVAAAD
trp2 SI(V/M)N/DA
trp3 FVFD(S/A)A(H/G)R
trp4 (V/G)(F/Y)LSKTR(A/T)RIC(M/G)R
trp5 S(?/T)(V/M)MH(A/D)MHA
EBK1-CT34 (V/M)(N/D/A)MA(N/D)S(A/Y)P(A/G)(G/M)(G)M
EBK1-CT61 (V/G)(Y/F)(V/G)(G/F/A)(N/V/A)(L/T/A)(S/G/?)
EBK1-CT73 "No sequence observed" was the result sent by
Harvard to the defendants.
trp1 DAVAAAD
trp2 FVFDSAHR
trp3 SIVNRA
trp4 VFLSKTRAR
trp5 STVMHAMHA
The
key conclusion is that the sequences
claimed to have been sequenced do not match those that Harvard says it actually
produced for Holloman and Kmiec.
Indeed,
Harvard actually stated this explicitly in their 2007 review of the work they
had performed for Kmiec:
“These sequences
are not consistent with the data we provided.”


The complete email is presented in the Harvard Amino Acid Sequences document.
The
Harvard Microchemistry Laboratory also made it clear that there was no potentially relevant
missing data, by writing to the Harvard General Counsel:
“...none of the sequence data we obtained agrees with the data they
claimed was from our lab.”
“I am confident that there is no other data”.

Therefore,
the five peptides Holloman and Kmiec claim in their paper, KCH194, and in
subsequent grants as having been sequenced at Harvard either were not those
actually isolated (trp 1, 2, and 5) or did not have the sequences claimed (trp
3, 4).
Back
to TOC.
Expert
Assessments of the Harvard Amino Acid Sequence Situation
Three
expert assessments were made of the data obtained from Harvard. The principal
issue was whether any of the amino acid sequences obtained by Kmiec and
Holloman from Harvard were from the Rec2 protein. Secondary issues dealt with
individual responsibility and the potential for penalties and correction of the
literature.
In
the primary assessment, it was determined that Harvard was correct in claiming that there was no connection
between the sequence data they had produced and what was claimed by Holloman
and Kmiec. The protein specified by the amino acid sequence produced by Harvard
was most likely from Ustilago maydis, but it was not Rec2:

As
specified in the expert's letter, the likely identity of protein from which
amino acids were obtained by Harvard is given at http://www.ncbi.nlm.nih.gov/protein/71013156.
That protein is not Rec2, but rather:
LOCUS
XP_758559
192 aa
linear PLN 25-APR-2006
DEFINITION hypothetical protein UM02412.1 [Ustilago
maydis 521].
ACCESSION XP_758559 XP_400027
By
comparison, Rec2 sequence is found at:
LOCUS
AAA64741
781 aa
linear PLN 04-APR-1995
DEFINITION REC2 protein [Ustilago
maydis]. ACCESSION AAA64741
(Kmiec
had also had a second protein sample, labeled "RecB" sequenced at
Harvard during the period of interest. Harvard produced this data during
discovery as well. The first expert was unable to find related sequence from
any organism in the databases she searched; however, it was determined that the
RecB amino acid sequences did not emanate from U. maydis Rec2.)
The
Thomas Jefferson University (TJU) protein facility, part of Kmiec’s former
employer and a co-defendant of Kmiec’s, did not provide any evidence regarding
whether they had performed any amino acid sequencing as claimed by Kmiec and
Holloman in KCH1994.
However,
as the claimed N-terminal sites purportedly sequenced at Thomas Jefferson
University no longer exist in the rec2-1
deletion mutant’s genome (see below), but the purported purified protein and
activity appear unchanged,
I have argued that there was no relationship between whatever might have been
sequenced at TJU and any Rec2 protein purified from its host fungus.
More
importantly, I have argued that examiners are entitled to draw an adverse
inference,
which in this case means that, as TJU and Holloman and Kmiec had good
reason to produce any evidence that "Rec1" (KSSEP) did have Rec2 amino acid sequences if
they had indeed found such at TJU, but did not do so despite substantial
opportunity during the investigation of this case, then it is reasonable to infer that no
such evidence exists.
Therefore,
not only have Holloman and Kmiec failed to provide any evidence that data
existed to support their claims that the amino acid sequence of the purported
“Rec1” strand exchange activity (KSSEP) was that of Rec2, but the evidence that
was obtained from Harvard University indicates that with a very high likelihood
the results claimed were based on falsified or fabricated data. (Whether the
misconduct is called falsification or fabrication might come down to the degree
of relation between the data obtained and the results claimed. In this case, as there appears to be no relationship to
the actual amino acid sequences, it is suggested that the data was fabricated,
i.e. essentially made up from whole cloth to meet the needs of the
researchers.)
Assessing
Responsibility For Falsified Data - Allegation 1
Given
that the amino acid sequence data appears with reasonable certainty to have
been, at a minimum, falsified, the
next question addressed by two of the experts was that of responsibility.
Kmiec
received the primary data from Harvard, and from what Rubin has stated,
produced the original manuscript which would become Kmiec et. al. 1994
("KCH1194"). It seems clear from Rubin's statements after having read
the Kmiec manuscript (see above) that a major point made by Kmiec was that
amino acid sequence of the Rec1 protein activity had purportedly been shown to
emanate from Rec2. Therefore, it does not seem likely that Holloman changed
truthfully reported amino acids sequences in the paper (i.e. that Kmiec
reported correctly), nor made major new claims about Rec1's identity with Rec2.
Thus, Kmiec would appear to have been a principal actor in
falsifying/fabricating this data.
It
remained, however, to assess the responsibility of Holloman, who stated in
writing during court proceedings that he had "relied upon the sequencing
information Kmiec provided".
But was this reliance reasonable?
As
noted in the Background and Basis for Allegations sections, Holloman knew
several issues had arisen with respect to Kmiec's work in science, including
much that had been performed with Holloman himself:
1) Kmiec’s Rec1 (KSSEP) work with Holloman from the
1980’s had never been replicated in any meaningful way by anyone else (other
than Kmiec) in Holloman’s or any other laboratory;
2) Holloman also knew that a serious controversy and
retraction of Kmiec’s work as a post-doctoral fellow had occurred. As Holloman
had written to Kmiec's post-doctoral advisor, Abraham Worcel, in the late
1980's regarding the TFIIIA issue:
“There has been
enough talk in the scientific community so that Eric’s results are looked at
with skepticism”.

(from information obtained by G.
Taubes).
3) Holloman also knew that on the very same KCH1994 paper
in which he and Kmiec claimed that “Rec1” (KSSEP) strand exchange activity had
the amino acid sequence of Rec2, two members of his laboratory, an M.D./Ph.D.
student (“B” is Rubin) and a post-doctoral fellow, insisted on removing their
names as authors because they did not find activity for Rec2 protein as claimed
by Kmiec. As noted above, the student told me:

4) Holloman also knew that he had published two papers
which showed genetic and biochemical data indicating that “Rec1” (KSSEP) strand
exchange activity was present in rec2 mutant cells but not rec1 mutants.
Indeed, Holloman referred to this situation as "paradoxical" in
KCH1994 itself:

return to DNA sequence falsification, below
5) It is further alleged here that Holloman knew that the
rec2-1 mutant had a large 5’ deletion in its rec2-1 gene that would have removed its active site. (See the
evidence obtained in the next section on DNA Sequence Falsification).
Nevertheless,
Holloman accepted Kmiec’s claims for "Rec1" (KSSEP) amino acid
sequence without verification. Indeed, it had been my experience that Holloman
had consistently argued against any of Kmiec’s work with him as being incorrect
despite the failure of numerous others in his laboratories to replicate it.
Rather, Holloman claimed to others that inability to reproduce Kmiec’s results
was the result of the inferior abilities of those trying to replicate, not any
possible wrong-doing. As noted by Taubes:

Above and beyond the
aforementioned red flags, Holloman also had a strong motive for wanting Rec1
(KSSEP) strand exchange activity to derive from the REC2 gene:
1) Holloman was aware that there was serious skepticism
about his work with Kmiec on the "Rec1" (KSSEP) protein, as he noted
in an NIH grant application:
2) The sequence of the REC1 gene had not shown any motifs known to be conserved among DNA
recombinases. Therefore, it seemed unlikely that the REC1 gene had encoded “Rec1” (KSSEP) strand exchange activity, (if
in fact “Rec1” (KSSEP) strand exchange activity had ever existed).
3) Holloman also knew at the time he published KCH1994,
that the REC2 gene sequence did have motifs that were consistent with being a DNA
recombinase. Ultimately, Holloman would be the principal beneficiary of large
amounts of NIH grant funding if he could justify and continue the “Rec1”
(KSSEP) work as associated with the real
REC2 gene.
Indeed,
it is important to note that it was Holloman who went to Kmiec and provided him with the Rec2 expression
construct when Holloman's own laboratory members could not obtain activity
which would connect what he and Kmiec had purported to be "Rec1"
strand exchange activity with Rec2 protein. As Holloman's graduate student
Rubin stated:
“So he [postdoctoral fellow Naoto Arai] came, and he tried to figure
out if the Rec2 protein was a strand exchange protein. He worked on it for,
well, almost two years. I'd say about two years. And he got the same results I did. Basically, we could never show this
did anything. ... what Bill did was
he sent my overpurification, Ok, so he sent my overexpressing strain to Eric's
[Defendant Kmiec’s] lab. Because, in my heart, I kind of believe what
happened was with him was, he just really wanted this result. To be a
strand exchange protein. And
I think what he did was he figured, well, these guys in my lab are not getting
this. So I am going to send it to Eric".
In
addition to wanting Rec2 to have strand exchange/transferase activity, Holloman
also had a major motive that Rec2 be the source of the strand exchange activity
he and Kmiec had ascribed to Rec1, i.e. especially once Holden et. al. had
published the REC1 gene sequence and
comments that it did not seem to be a strand exchange protein (see above).
Kmiec,
as Holloman's collaborator on the Rec1 claims, which had also proven to be
irreproducible by others, especially in Holloman's own lab, shared the same
motives and goals. It remains remarkable that Holloman not once but twice
apparently ignored the negative findings of multiple of his own laboratory
members in favor of those from Kmiec, whose reputation after the TFIIIA
retractions would have raised at least some concern about his production of
remarkable results. As noted above, Holloman explicitly had written just this
himself: "There has been enough talk in the scientific community, so that
Eric's results are looked at with skepticism."
Therefore,
Holloman's thinking about the perception
of Kmiec was rational. Yet he himself refused to apply any skepticism at
all; he stated in the qui tam case
that he simply "relied upon" what Kmiec provided to him. An obvious reason for doing so was that
Holloman got just the results he wanted. Hence in the legal case we
alleged that the relationship between Kmiec and Holloman could be seen as a
conspiracy.
The
standard by which one is determined to have had "knowledge" of a
fraud, as defined by the applicable U.S. federal law (31 U.S.C. 3729-3733), and
consistent with the legal principles of scienter, is the following:

Therefore,
the question arose as to whether it was more likely than not (greater than 50%
likelihood), that with respect to accepting Kmiec’s claims that “Rec1” (KSSEP)
had the amino acid sequence of Rec2, Holloman was acting with reckless
disregard for the truth or in deliberate ignorance.
Subsequently,
two additional molecular biologists reviewed the information obtained from
discovery. With
respect to the issue as to whether misconduct had likely occurred, they
commented as follows:
1.
Do you believe that K acted intentionally in reporting that Harvard
Microchemistry had obtained R2 amino acid sequence when [they] did not do so?
"Yes." ExpRev-1
"Yes. In both my experience as a
primary collector of data and now more recently as a lab chief, this type of
error is not possible. The process has too many checks and balances, preventing
an unintended mistake." ExpRev-2.
2.
Given what H knew of K’s history of irreproducible and retracted results, and
the “unexplained” “paradox” (in H’s words) that R1 strand exchange activity
should derive from the R2 gene, was it reasonable for H to have “relied upon”
K, for such a vital result (putative R1 strand exchange activity had the amino
acid sequence of R2)?
"Clearly
unreasonable." ExpRev-1
"It was not reasonable. This in many ways is a critical
issue. There is absolutely no
reason in my opinion to have an expectation of certainty regarding K's work
given the other available information." ExpRev-2.
4.
Do you believe, given all the evidence, that it was more likely than not that H
was acting in “deliberate ignorance” or with “reckless disregard for the truth”
by accepting K’s claims about R2 sequence from putative R1 strand exchange
activity?
"Yes. More
likely than not H knew the truth, but can you prove this?"
ExpRev-1
"I can't help but believe from my
perspective as the Principal Investigator of a laboratory that it was
deliberate ignorance. He knew full
well that things were not appropriate." ExpRev-2
5.
Knowing now that Harvard Microchemistry does not support claims made about work
attributed to it in the paper KCH1994, now 16 years old, what do you believe
should be done?
a)
Should the journal retract
the paper?
b)
Should K be subject to any
penalty? Should H? If so,
what?
c)
Should the NIH take such information into account in considering future
funding
for these
researchers?
"I believe
that the journals should retract this data despite how long ago it was
published. In fact they should provide space for a detailed account of the
story. K should be tried for falsifying data. H should lose his funding. I do
not know if there is any mechanism to recover past misused MIH funds."
ExpRev-1
a) "YES, setting the
record straight is necessary and appropriate even given the length of the
intervening years."
b)
"YES. Both K and H have engaged in misconduct.
H is far more at fault as the supervisor and the person ultimately in
charge. ..."
c)
" They should be barred from
receiving NIH funding in the future. ..."
Back
to TOC.
II. DNA Sequence Data Falsification - Second Allegation
It
was Holden et. al., in a paper in Current Genetics (20:145-150) in 1991, who
first exposed a problem that had arisen from sequencing the DNA of U. maydis' recombination genes. As they
stated:
“We have also cloned the gene in order to determine if
indeed REC1 encodes the
transferase protein [KSSEP] or, for example, a protein which regulates
expression of the transferase and perhaps other genes involved in recombination
and repair.
“We have found that the amino acid sequence deduced for
the Rec1 protein is not compatible with the molecular size of the transferase
[KSSEP] (Kmiec and Holloman, 1982). It also does not contain any of the
conserved ATP- or DNA- binding motifs, although a nuclear localization
signal may be present (Holden et. al., 1989 b)”. (Emphasis added.)
Therefore,
nucleotide sequence of the REC1
gene indicated that, contrary to Holloman's hopes, the REC1 gene did not have a sequence comparable to other known
recombinases. If it had, he would have claimed justification of his earlier
KSSEP results with Kmiec.
Instead,
the difference put Holloman in a precarious position. Any new grants could be
attacked on the basis that his claimed "Rec1" strand exchange
activity (KSSEP) would have been unlikely to have had the size and activities
originally claimed by him and Kmiec.
A
much more obvious explanation, given Kmiec's subsequent history of retracted
experiments and failure of Holloman's own laboratory personnel to extend the
Rec1 strand exchange work, would be that the original Rec1 (KSSEP) claims were
also false. Therefore, Holloman faced the likelihood that any critics among his
federal reviewers would again reject his grant applications.
Motivation and
Importance of Rec1 becoming Rec2
But,
"as fortune would have it", as Holloman stated in a federal grant
application, the sequence of the REC2
gene did have a sequence compatible with activities they had described for
“Rec1” strand exchange activity (KSSEP).
The
associated document "REC2 and rec2-1 technical information" includes diagrams of the
various forms of REC2 gene and Rec2
protein discussed in this section.
In
addition to the "paradoxical" biochemistry and genetics from "earlier published results
[being] virtually opposite to expectations", i.e.
with Rec1 having emanated from the REC2
gene, as noted by Holloman, there were some additional issues related to the
sizes of the expected protein products.
The
most important size issue, to me, was that the purified "Rec1" strand
exchange activity (KSSEP) was always obtained as a pair
of approximately 70 kd peptides (see Background above). However, not only was
KSSEP activity found in the rec2-1 mutants, but so were the 70 kd peptides (see
Western blot for an example in the "REC2
and rec2-1 technical
information" document).
The
problem with this is that it was already known by the time KCH1994 was written
that rec2-1 had been formed by a
large deletion of the 5' end of the REC2
gene, e.g. as shown by Southern blots and subsequently DNA sequence published
by Holloman in the Rubin et. al., 1994 paper.
However, as noted in the Basis for Allegations section above, Holloman had
apparently removed information specifying the 5' end (of the open reading
frame) and exact deletion points of the rec2-1
deletion from the Rubin et. al. 1994 paper.
Rubin: You know
what is funny is, that somehow the, I actually did notate that the
rec2-1 mutant, that on this figure originally, Figure 1, and uh, in the legend,
5 the description, I am looking at my thesis right now, and it was the same
figure, and it is in there. Bauchwitz:
Oh, it is? Rubin: Bill
must have taken it out for some reason.” (Sec. Decl., par. 28.)
The
proteolytic digestion sites claimed in KCH1994, which supposedly reduce the
larger Rec2 protein to 70 kd peptides, fall within the rec2-1 genomic deletion. Therefore, it would have been a remarkable
coincidence that the products of proteolysis would produce a peptide pair of
effectively identical size as a deletion product of the same gene -
particularly when the digestion sites had been removed by that deletion!
Important
evidence from the rec2-1 gene sequence
Even
if a presumptive rec2-1 protein product were expected to be much smaller in
size (56.5 kD predicted) than the 70 kD pair observed in the rec2-1 cells, it
would remain unknown whether there was any sequence upstream of rec2-1 which would extend the open
reading frame to make a longer composite protein.
Of
even greater importance than peptide sizes, if the rec2-1
mutant allele were not able to produce an active protein, then normal levels of
such activity could not have been isolated from rec2-1 cells by Kmiec and Holloman, contrary to their claims
in two prior publications (Cell 29: 367-374, 1982; Cold Spring Harbor Symposia
on Quantitative Biology, Vol. XLIX, pp. 669-673, 1984).
Examination
of the rec2-1 DNA sequence upstream
of and across its deletion point should have allowed an assessment of possible
size and especially activity issues, since the active site of the protein had
been determined. Although Holloman's graduate student, Rubin, had told me that
he had sequenced the rec2-1 allele, I
could not find any specification of the relevant upstream sequence, not only in
his 1994 paper (as he had noted in surprise, see above), but also not in his
thesis, nor in the federal Genbank.
Therefore,
in 1997 I produced my own DNA sequence of the rec2-1 deletion breakpoint and I also extended the upstream
sequence of REC2. I deposited those
sequences in Genbank and made them available to the public in 1999. (Some of
the raw data I obtained is shown in the "REC2 and rec2-1 technical
information" document.)
...
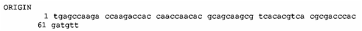
The DNA sequence results I obtained were
consistent with what Rubin had told me he had found while he had worked in
Holloman's laboratory. These results appeared to be a significant piece of
evidence against Holloman's claim that Rec1 had actually emanated from the REC2 gene, since the results were now
not only paradoxical and opposite of expectations, but incompatible - there
should not have been any Rec1 activity found in rec2-1 cells if those cells
were not capable of producing any active recombinase.
Just
how definitive my rec2-1 DNA sequence
was with respect to being put on notice of fraud was discussed at length by the
Court in this case (Memorandum Opinion re Summary Judgment, doc. 116). It should be noted that U.S. qui tam law (31 U.S.C. 3729 - 3733) requires that an actual false
statement be made with respect to funding from the federal government. In other
words, just publishing a false statement is not enough to trigger the laws
under which this case was pursued. The false statements must be made as part of a
false claim for payment or certification such that the taxpayers are
defrauded of their funds. Holloman never made an explicit statement about the rec2-1 DNA sequence, at least not one
that I knew of, until his 2001 publication with Kojic et. al. (see detailed in
the following section). The grant relevant to the rec2-1 false statements was first funded in 2002.
Nevertheless
the Court stated, " Bauchwitz knew the facts material to this second
category of fraud as early as February of 1995, when he spoke with Rubin for a
second time, and later, in 1999, when he published the results of his own rec2-1 sequencing."
Leaving
aside that there are many significant inaccuracies in the judicial opinion as
to the facts that had been presented,
the question is whether I was really in any position to file a lawsuit based on
the inconsistency of the rec2-1 DNA
sequence result with the Hollomans' claims that Rec1 had Rec2 amino acid
sequence. Was that information really enough that the clock began to tick on my
ability to take legal action to recover federal funds related to the first
allegation of fraud? What I did early on was suggest to the relevant federal
agency, ORI, that they investigate. They apparently did not do so.
As was stated in the court record on this point:
"The only question with
respect to alleged frauds 1 and 2 would seem to be whether I had any duty in 1995
- long before I even knew what a qui tam
action was, or had even considered whether defendants might seek future federal grant funding based on
the False Claims 1 and 2 – to act as a perpetual investigator and monitor
of future grant applications by the defendants, based solely on my skepticism
about the results reported in the 1994 Paper. I respectfully submit that that this
cannot be the case. To the
contrary, I believe that in 1995 I went above and beyond the call of duty by
notifying ORI of continuing suspicious circumstances that warranted investigation,
and subsequently developing my own rec2-1 sequence, when nothing by Rubin could
be found." Sec Decl. par. 32.
I
do not agree that such inconsistent results are as definitive with respect to
knowledge of fraud as the Court seems to portray them. As I argued (see Basis
for the Allegations section, above), had I not known of the prior history of
concerns about the work of these researchers, I might have assumed that
technical errors had occurred to
produce the inconsistent results. Indeed, this is precisely what was stated in
the court record:
"Rubin stated, he was “very
skeptical about all this”.
(emphasis added). I too was skeptical about the results reported in the
1994 Paper, but note that elevated skepticism about the results
reported in a published research paper is not the equivalent of knowledge
that a fraud had occurred, or even evidence of such. ... It was only in
the context of their prior pattern of dishonest behavior, which preceded and
was separate and distinct in nature from the current information I was hearing
for the first time, that I felt an investigation by ORI was warranted in
1995." Sec Decl. par.s 16 and 17.
Regardless
of just how much notice my rec2-1 DNA
gave me, the Court clearly considered this sequence and related information
(e.g. Holloman not presenting his own) highly
material to understanding the case; the same has been true for scientific
reviewers to date.
Holloman
did eventually publish a specific claim about the rec2-1 DNA sequence, at which point, in conjunction with the
presumptively false statement appearing in relation to funding from the U.S.
government, it is without doubt that the statute of limitations would begin, at
least for the second alleged fraud.
Publication of rec2-1 sequence claim by
Holloman
While
I was assisting journalist Gary Taubes, he brought to my attention an article
published by Holloman in 2001 in which Holloman claimed that the rec2-1 sequence did
have an upstream ATG which would have allowed a rec2-1 peptide to retain the
recombinase active site:
"Inspection
of the rec2-1 DNA sequence on
both sides of the deletion indicated that a novel ORF could be generated
through conjunction of the flanking sequences. This ORF would be
predicted to encode a 613 amino acid [Rec2] protein variant with a novel
19-residue leader sequence derived from upstream of the deletion in lieu of
the N-terminal 187-residue sequence of the wild-type [Rec2] protein.” (Kojic et. al., 2001.)
Holloman
was claiming that an ATG DNA sequence exists upstream of the rec2-1 deletion site in a position which would make it
reasonable to assume that a protein could be produced from rec2-1.
This
despite testimony that data Holloman received from his graduate student, Brian
Rubin, did not indicate such a sequence, and despite public data (GenBank
AF027108 of 1999) also to the contrary.
Of
particular note, Holloman never explicitly showed the relevant sequence or
published it in the Genbank. He only made statements and showed schematics in
the paper.
This
naturally raised the question as to whether Holloman and his new authors
actually had rec2-1 DNA sequence to
support his claims.
I
summarized the situation in a disclosure to the U.S. government (ORI and
Department of Justice) as follows:
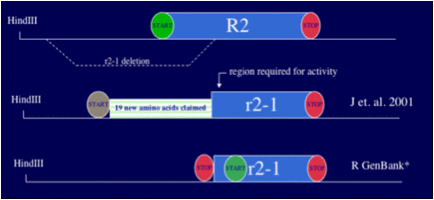
(*)
rec2-1
should not have been able to produce the “Rec1” (KSSEP) protein activity.
To summarize the
motivations for the 2001 rec2-1 DNA
sequence claim, if
there were no ATG start codon, and hence no methionine which almost all
proteins have as the first amino acid, the likelihood of a protein from rec2-1 would be so remote that without further evidence it
would not be proposed as a reasonable explanation of activity from the mutant
gene rec2-1. Therefore,
claiming that ATG was present made Holloman's explicit and implied claims of
potential activity from rec2-1
seem to have been plausible.
In
other words, while it would be possible to use “hand waving” to explain why rec1
mutants did not show expected KSSEP protein activity (a negative result), it
would be much more difficult to explain why purified KSSEP activity would be
present in rec2 mutant cells (a positive, affirmative result) if those
cells did not have a rec2
gene capable of producing an active Rec2 protein.
Back
to TOC.
New Information about the rec2-1 DNA sequence
What
follows is new information that was obtained by subpoena during discovery in
2010 regarding the rec2-1
sequencing issue. Holloman was required to respond to written questions under
oath and provide relevant documents. However, Holloman’s graduate student,
Rubin, could not be questioned; all that is available are his statements to me
in 1994 and 1995.
There was no other rec2-1 breakpoint sequence produced by
Holloman other than by his graduate student, Rubin
Holloman,
in his sworn response claimed that he relied upon the rec2-1 DNA sequence numbering in his graduate student Rubin’s
thesis for his claims in his 2001 paper.
When
asked for the identity of anyone else involved in the rec2-1 sequence at issue, Holloman did not specify anyone
else, but again referred only to the sequence produced by his graduate student,
Rubin.
When
asked, Holloman also specified no other source for the rec2-1 sequence he claimed in his 2001 paper other than Rubin’s.
Therefore,
there appears to have been no rec2-1 DNA sequence performed by Holloman’s laboratory after
Rubin’s work, e.g. nothing by Kojic or Thompson, the other authors
of 2001 paper.
The DNA sequence position numbers claimed to have been
relied upon by Holloman could not have produced the results he published.
The
following analysis of the relevant new evidence is technical in nature. For
those who do not have experience in assessing DNA open reading frames, this
material (shaded in gray) can be skipped. For those who have some experience in
molecular biology, the original analysis submitted to the Court (attached, Reviewer
Documents, 3a, on the CD) has been supplemented here with some explanatory
material, which should make it more accessible to those who are less
experienced. A summary and reviews by experts in molecular biology are provided
in the subsequent section.
The specific
sequences at issue
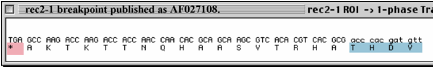
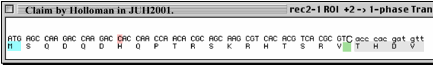
Notes:
The extra “TC” at the breakpoint claimed by H is shown at the green box in second
figure, above. These two nucleotides originate from just beyond the end of 5’
side of the deletion breakpoint.
In
a written response to questions, Holloman claimed that he had relied upon a DNA
sequence from Rubin's thesis (here the "B" stands for "Rubin"):

However, Rubin’s thesis does not show the sequence
stated by Holloman, above, nor does it even imply the “TC” at the end of the 5’
deletion breakpoint. (See the "REC2
and rec2-1 technical
information"
document for details on the numbering scheme used in Fig. 1.3 of Rubin’s
Thesis).
Upon
production of written documents, it was revealed that the information that
Holloman cited was present as marginalia
on a REC2 DNA sequence summary
document.
On
that sequence document, dated June 17, 1991, which was found among other
sequencing records apparently produced by Holloman’s graduate student Rubin,
has been drawn in a very heavy black line a demarcation of what exactly
corresponds to the recr2-1 deletion breakpoint sequence as it appears in the Genbank
entry AF027108, produced by me. (See the "REC2 and rec2-1 technical information" file for relevant excerpts of the
documents produced; the complete set of relevant documents can be found on the
CD in Reviewer Documents/3c.)
This
strongly indicated that Rubin, the only other person known from this case to
have sequenced the rec2-1 deletion
breakpoint, had obtained the same sequencing results as I had.
The
next question was whether there was in fact some dispute as to the presence of
an ATG start codon upstream of the rec2-1
deletion breakpoint. Such a start codon would have been required to retain the
Rec2 active site in any rec2-1 protein.
Open reading frame (ORF) analysis is used to predict peptides
which might be produced by a segment of DNA. In essence, the process looks for
known start and stop signals which control where protein synthesis machinery
begins to read the sequence (via its RNA intermediate form) and when it stops
synthesis. As with the ATG start codon in DNA, stop codons are also present.
This part of the analysis is relatively simple - look for known start and stop
triplets of DNA in the gene sequence of interest.
An
extra level of complexity is introduced because there are three potential
reading frames in each direction of a double-stranded DNA. For example, we know
that the amino acid sequence THDV (shown in the rec2-1 breakpoint sequence
figure in the REC2 and rec2 technical
information technical file, found the CD)
must be present beyond the 3' deletion breakpoint in any Rec2 or rec2-1
protein. Those amino acids are coded by the DNA sequence triplets: acc cac gat
gtt. Therefore, those amino acid and associated DNA sequences anchor the
reading frame that must be used.
Then,
one can work towards the upstream, DNA triplet by DNA triplet, to look for an
ATG start codon. To be in-frame means that the ATG corresponds to the position
expected for a triplet, rather than being out-of-frame by overlapping two
triplets. For example, ATG CCC acc cac gat gtt would put the ATG "in-frame"
with the amino acids encoded by acc, while ATG CC acc cac gat gtt would have it
out-of-frame; the actual result would be xAT GCC acc cac gat gtt, for which x
represented the next DNA nucleotide upstream of the AT. Performing this
ORF analysis is aided by the use of computer software.
The
final point to be considered with the discovered evidence is that Holloman
never actually presented Rubin's actual rec2-1
open reading frame as a singular, unbroken string, despite subpoenas for the
information. He
only showed it within the context of where it existed within the wild-type REC2 gene sequence.
Nevertheless,
it is relatively easy to put the two pieces of the Holloman sequence that
correspond to rec2-1 together. The
result shows precisely what was published in the Genbank as AF027108, and as
shown in the top portion of the preceding "Specific sequences at
issue" figure. This sequence is not what was claimed in the 2001 Holloman
paper (Kojic et. al., 2001) nor by Holloman during the court proceedings (see
preceding figure, "B's thesis states").
While
it is essentially certain that no one would make a specific sequence claim
about an open reading frame without actually assembling one as a single,
continuous sequence, i.e. in a manner sufficiently specific that the sequence
would be appropriate to enter into a database such as Genbank, it might be
possible to use numeric shorthand based on relative DNA sequence positions to
describe such an open reading frame in a way that would satisfy some research
journal reviewers or editors. The basis for so doing would be that they would trust the author to have carefully
checked his numbers. But responsible reviewers would not have accepted that an
author would have produced a claimed open reading frame without actually assembling it. In other words, pretending to rely
on someone else's numbered sequence positions would not be appropriate.
Holloman
presented such sequence numbers in two instances in this case. First, as shown
above, he wrote: "A = +1 (-231-+563)". Second, in the DNA sequence
that Holloman released are numbers written in hand among the sequences and in
particular, in the margins ("marginalia").
"A
= +1" is a way of saying that the first nucleotide of the wild-type REC2 DNA sequence, which starts with the
usual ATG, will be numbered "+1". This is standard numbering for such
sequences. Indeed, it is also shown on the corresponding DNA sequence document
he released, in which the REC2 ATG
start codon has a handwritten box around it and a "+1 Met" with a
rightward pointing arrow above it:

Furthermore,
the "(-231-+563)" numbering also appears again in the margin of the
DNA sequence document as a handwritten: "Deletion = -231 to +563". We
were not told who the author of those marginalia was, nor when they were made.
The
-231 to +563 numbering represents the position of the deletion endpoints
relative to the A being +1. Establishing these numbers can be done by a
relatively straightforward counting of the DNA nucloetides between the two ends
of the deletion, for example as follows:

The
figure immediately above shows that the DNA sequence software itself applies
numbers to the nucleotide bases based on the first base in the file being +1.
Thus, the numbers "210" and "280" can be seen in the right
hand margin; these correspond to nucleotides at the end of each line.
Much
of the following discussion was presented in the case document (Plaintiff's Third Interrogatory Response of
March 25, 2010, question 8, which was presented to expert reviewers, with
one added figure, as found on the CD as file 3a_r2-1_seq_new_data_analysis_alt_2010.pdf).
Importantly,
above the “T” immediately following the end of the heavy black line
corresponding to the 5’ end of the rec2-1
DNA sequence, has been written the number “238”. This number correctly
identifies the position within the sequence file of the “T” above which it is
present. This indicates that whoever wrote "238" could count
correctly either up from 211 at the start of the line, or back from 280 at the
end of the line.
Similarly,
if one reexamines the previous figure that shows the boxed "ATG" with
"A met" above it, it is apparent that there is a "490" at
the end of the same line. Furthermore, in light handwriting a “470” has been
written in by hand under the “C” which precedes the ATG. The “A” defined as
“+1”, is clearly at position 471 of the sequence file, so again, whoever wrote
the "470" could perform this simple counting.
Finally,
the discovery document showing the 3' end of the rec2-1 deletion (see following), indicates that the rec2-1 deletion ends at position 1034 of the DNA sequence file
and also correctly shows the indicated HincII marker as being at position 1091.
Once again, these numbers are easily generated by counting from the end of each
line.

Therefore,
all of the aforementioned numbers drawn within the DNA sequence are correct and demonstrate that whoever
wrote them could count using the numbering provided by the DNA sequence
program. (Note that numbers in blue are not part of the original documents but
were added to facilitate counting by reviewers.)
Notable
Discrepancies
The
marginalia that claim "Deletion = -213 to +563", however, appear to
be inconsistent with the data presented and the aforementioned markings.
First,
if the ATG is at position 471 and the 5’ deletion begins at 238, as indicated
in the documents, then the deletion would begin at -233, not -231.
Furthermore,
if the 3’ end of the deletion is at 1034, then the deletion would end at +564,
not +563.
The
most direct way to establish these facts would be to count every nucleotide on
the DNA sequence documents, e.g. as they appear in their complete form (CD file
3b in folder Reviewer Documents).
Molecular
biologists, and those with comparable skills, could use specialized software
that could adjust the numbering scheme, e.g. to set the A = +1, or otherwise to
produce a count of the nucleotides between two points.
A
third method would be to use some simple math involving the endpoint numbers,
i.e. subtraction.
If the person who
wrote the marginalia thought that it was appropriate to simply subtract the
sequence file positions to obtain the deletion point positions, then they would
have obtained:
For
the deletion end point,
1034
– 471 = 563, as stated, but,
For
the deletion start point,
471 – 238 = 233,
contrary to the marginalia claiming 231.
Above
and beyond the aforementioned problem, the number “+563” is not a correct
determination of the actual sequence shown.
When
one subtracts, one is determining a difference, e.g. the number of steps
between two points. For example, if a person wanted to count the 5 fingers on
his hand, then he would not subtract 5 – 1, as this produces 4, which is
not the number of fingers. To go from 1 to 5, a person would need to take four
steps, but it is necessary to add back “1” to get the actual number of fingers (or
nucleotides) separating two points.
The
problem does not occur when going backwards in such a sequence file because it
contains a “-1” which gets included in the difference, i.e. it obviates the
need to add back a 1. So the negative numbers would be correct.
The
net result is that +563 is NOT the end of the deleted sequence and could not have been relied upon to produce the rec2-1 sequence claimed by Holloman in the answer he made on
March 8, 2010 (see preceding).
Clearly,
had Holloman relied upon the number +563, with “A” of ATG = +1 as he claimed,
he would have found from any REC2
sequence that an additional “A” would have been present at the start of the 3’ rec2-1 breakpoint sequence.
Therefore,
I concluded that Holloman did NOT rely upon this “erroneous” number +563 to
produce the sequence he claimed, because if he had done so, an open reading
frame analysis would have shown only six amino acids possible following the 3'
end of the deletion breakpoint, i.e. following the 19 novel amino acids claimed
from the 5’ end of the rec2-1
sequence in Kojic et. al., 2001:

The red asterisks here are in-frame stop codons. The
blue "M" is the start of the rec2-1 amino acid sequence claimed in
Kojic et. al., 2001.
Thus,
there is no connection between the erroneous numbers supposedly
relied upon and
any rec2-1 DNA sequence claimed by Holloman, either in Kojic et.
al., 2001, or in his interrogatory response.
Summary of the
immediately preceding reviewed evidence for the allegation of DNA sequence
falsification
In
short, the above reviewed evidence shows that Holloman could not have used the
numbers “ATG start A = +1 (-231 -- +563)” to produce the rec2-1 sequence as he claimed in his interrogatory answer
number three of March 8, 2010 and as he showed in his publication Kojic,
Thompson, and Holloman, 2001. Whoever produced the number +563 apparently
misunderstood that the process of subtraction could not be used without an
adjustment (adding one).
It is notable, and
we make adverse inference from, the fact that despite the request made
in writing during court discovery, Holloman failed to produce the actual open reading
frame sequence he purportedly must have produced if he had in fact relied on
the numbers he provided, as he stated under penalty of perjury. Holloman never
presented an actual rec2-1 ORF that
was produced by him or any author of Kojic et. al., 2001.
Instead,
the evidence
indicates that Holloman simply chose the sequence he wished to obtain and then
presented it without any attempt to determine if it was consistent with what he
knew was the true REC2
sequence, much less with an open reading frame assessment of the rec2-1 mutation.
Back
to TOC.
Many Missed Opportunities to Present rec2-1 ATG Claims
or Actual DNA Sequence
Despite
Holloman’s claim for the rec2-1
sequence in Kojic et. al., 2001 ("JUH2001"), in at least one half
dozen instances in grants, papers, theses, and patents in discussing this issue
up to 2001, Holloman did not ever make a claim for the existence of the
upstream ATG, despite his clear recognition by his written statements of its
relevance and importance.
It
was not until after the publication of the Genbank rec2-1
sequence in 1999 that Holloman claimed a contrary finding in the Kojic et. al.,
2001 paper.
However,
even there, he only did so through textual statements and schematics. Holloman
did not present an actual rec2-1
DNA sequence nor, apparently, to this day has he placed such in the Genbank or
any other comparable public database.
Examples of “missed opportunities” to
disclose the vital upstream ATG
The importance of an
alternative rec2-1
translation start site was already noted in the Discussion of Kmiec, Cole,
Holloman, 1994:

If
Holloman had believed that an alternative start site for translation existed
upstream of critical amino acids in the rec2-1
allele such that “certain biochemical activities”, e.g. as associated with a
DNA strand exchange recombinase, could have been produced, he most definitely
would have been expected to mention it here.
Yet
he did not, even though he had the rec2-1
sequence produced by his graduate student, Rubin.
The
first mention of such ATG was not until publication of Kojic, Thompson, and
Holloman in 2001, and according to court testimony by Holloman, it was not
based on any new rec2-1
DNA sequence.
Even
Holloman’s graduate student, Bennett, who followed Rubin in the laboratory, did
not mention such an ATG in his 2001 thesis when reviewing the rec2-1 allele:
“The rec2-1 mutant is the result of a deletion extending
600 bp into the ORF from the ATG codon and including 200 bp of upstream sequence
(Rubin et. al., 1994). However, a truncated mRNA is observed in northern blot
hybridization using REC2 sequence as a probe for detection.” (Bennett Thesis, p.38.)
Bennett's
mention of mRNA was apparently intended to hint that protein might be produced.
Yet only the Rubin rec2-1
sequence is cited and no upstream ATG mentioned.
The
Bennett thesis was dated May 2001, and like Rubin’s thesis, apparently signed
by Holloman. It seems apparent there was no general knowledge or discussion in
the Holloman laboratory about an upstream ATG, even at this late date,
contemporaneous with the Kojic et. al., 2001 paper which proclaimed the
upstream ATG.
The
Office of Research Integrity made similar note of this situation, e.g. in a
January 12, 2005 letter to the Department of Justice:

and no mention of an upstream ATG in a relevant grant:
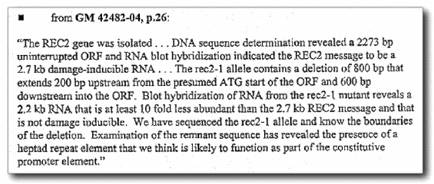
Back
to TOC.
Expert
reviews of the rec2-1 DNA sequence evidence
The
claims I made above regarding the rec2-1
DNA sequence were assessed by the two molecular biologists who reviewed the
material obtained during discovery in the case. Their complete reviews are
attached. The reviewers were not given the actual identities of the defendants
or others involved; the name codes used were the same as provided to the
federal government. In particular, I am "R", Holloman is "H",
and Kmiec is "K". The following are relevant excerpts of their
conclusions:
Does
the sequence data presented by R support his contention that the first ATG
initiation codon (following an in-frame stop codon) in the r2-1 DNA sequence is located downstream of
the r2-1 deletion breakpoint?
Yes. ExpRev-1
YES ExpRev-2
Does
the evidence indicate that B’s r2-1 breakpoint sequence agrees with that
published by R in Genbank?
Yes ExpRev-1
YES ExpRev-2
Does
the evidence provided by H support “a 613 amino acid [R2] protein variant with
a novel 19-residue leader sequence derived from upstream of the deletion” as
claimed in JUH2001?
No ExpRev-1
NO ExpRev-2
Would
H’s claim to have relied on the sequence numbers “-231 to +563” be consistent
with the 613 amino acid open reading frame he published in JUH2001? If so, how?
No ExpRev-1
NO ExpRev-2
Do
you believe it is more likely that not that H was acting in “deliberate
ignorance” or “with reckless disregard for the truth”, as defined by law (see
Introductory Note, above), in the manner in which he claims to have relied upon
sequence numbers for r2-1 findings stated in JUH2001?
H has tried to
construct a logical progression that can account for his and K's original bad
data. I do believe that data was fabricated by H and K. ExpRev-1
This is a tough call on this issue but
nonetheless it is one of these two conclusions. That is, taken as a whole I would have
said "deliberate ignorance". However, on this specific issue the
evidence would also be consistent “with reckless disregard for the truth”. ExpRev-2
Do
you believe that it is more likely than not that H made an innocent error or
was acting by incompetence in his claims about finding an ORF for r2-1 as described in JUH2001? If so, why?
I do not believe
that these mistakes were innocent. ExpRev-1
It is more likely that H was
deliberately making things up given the collective observations presented. In
either case, I would argue against an innocent error. ExpRev-2
Based
upon the information presented here, do you believe that it is more likely than
not that H falsified his r2-1 data claims in JUH2001?
Yes ExpRev-1
YES
ExpRev-2
If
you believe it was more likely than not that H was not acting by incompetence or innocent
“error” in making sequence claims for r2-1 in JUH2001:
a)
Should the journal retract the paper?
Yes ExpRev-1
YES
ExpRev-2
b)
Should H be subject to any
penalty? If so, what?
At least his grants
should be revoked. ExpRev-1
YES. H again appears to have engaged in misconduct.
That is, in my mind he is criminally at fault
- there is no excuse other than self-interested, self-motivated, self-indulgent
behavior.
ExpRev-2
c)
Should the NIH take such information into account in considering future
funding for this researcher?
Absolutely. ExpRev-1
He should be barred from receiving NIH
funding in the future. ExpRev-2
If
you were a member of an NIH grant review committee and learned there of these
facts concerning r2-1 sequence claims, regardless of
judgment of whether deliberate, would this have had a negative impact on your
scoring of the grant?
Yes. ExpRev-1
YES
ExpRev-2
Back to TOC.
III. Protein Activity Data Falsification - Third
Allegation
Due to
restrictions placed on the time for which we were allowed to perform discovery
(see document, "Case History"),
the only new evidence that I was able to obtain relating to the third
allegation of fraud came from Freedom of Information Act (FOIA) requests for
grant documents. Prior to filing the case, I had also obtained grants through
the FOIA, as well as publicly available graduate student theses. As the new
information obtained by FOIA did not require a technical analysis, it was not
reviewed by experts.
Nevertheless,
the FOIA grant documents, as well as the theses of two of Holloman's graduate
students, were very revealing with respect to the third allegation in
particular, and the entire alleged Rec1 is Rec2 fraud in general. Therefore,
information obtained related to the question of what was known about purified
Rec2 activity, both in 1994 as relevant to this case, and throughout the period
relevant to the grant documents (through 2007), is presented below.
To review from
the Basis for Allegations section, Holloman's graduate student, Brian Rubin,
had indicated to me that he was of the belief that data produced by him had
been used by Holloman in the 1994 Kmiec, Cole, Holloman paper to imply Rec2
activity, when in fact not only was his data from an inactive preparation, but all
preparations of such protein produced in the Holloman laboratory by him and
postdoctoral fellow Naoto Arai had been inactive.
On
December 27, 1994, Holloman's graduate student, Brian Rubin, made the following
comments:
Rubin: “I think of him as just a
guy that develops stories in his office and then comes into the lab and says
produce the data that fit my stories. And in fact in that paper, there are figures
straight from my thesis that have totally nothing to do, really, with what was
published.”
Bauchwitz: “What do you mean by
that?”
Rubin: “Well like the protein, he
[Defendant Holloman] used my purification gel and Western showing the anti-Rec2
antisera binds this protein.”
Bauchwitz: “Yeah.”
Rubin: “And then he claims that this protein
is active. And he would rationalize it by saying ‘Well, it's
just a better looking gel than Eric's [Defendant Kmiec]’ Of course, we are
using the same strain, but that prep wasn't active.
As Rubin also stated in that conversation:
Rubin: “So he [postdoctoral fellow Naoto Arai] came, and he tried to
figure out if the Rec2 protein was a strand exchange protein. He worked on it
for, well, almost two years. I'd say about two years. And he got the same results I did. Basically, we could never show this
did anything. And we purified numerous helicases from Ustilago and E. coli, and
basically we were looking for a DNA dependent ATPase. We tried to follow,
that was sort of our base assay, was DNA dependent ATPase. We also did
extremis. We tried strand exchange on
our purified preps, but we never got it to do anything. …
Therefore,
it was alleged that Holloman falsified Rec2 activity data presented in Kmiec,
Cole, Holloman, 1994, according to the information provided by his former
graduate student, Rubin.
In addition to
the specific issue of data characterization, more generally with respect to the
"Rec1 is Rec2" protein activity allegations I argued:
"Holloman’s reliance on irreproducible data purportedly derived by Kmiec,
and his
disregard of the data produced by researchers in his own laboratory,
which demonstrated no Rec2 activity at
the time of the publication and grant submission, demonstrate that Holloman
acted, at a minimum, with reckless indifference or in deliberate disregard of
the truth, given Holloman’s awareness that Kmiec had a serious history of
reproducibility issues: 1) his Rec1 protein activity work had not been notably
reproduced by those inside or outside Holloman’s laboratory (excepting
subsequently by Kmiec), and 2) Kmiec’s work as a postdoctoral fellow with
Abraham Worcel had been publicly retracted." (First Decl. par. 42, emphasis added.)
In response to
the third allegation, and improperly but repeatedly ascribed to all the
allegations, Holloman's attorneys used a publication of Holloman's in 2001 that
purported to show Rec2 protein purified from E. coli as having "Rec1"-like transferase activities (Bennett and Holloman, 2001), as a
defense against claims that he had acted inappropriately in trusting Kmiec over
those in his laboratory. I termed this the "Bennett Defense".
The following are excerpts from a discussion of this issue in the Sec Decl.
"The
Bennett publication of 2001 has no direct bearing on any of the allegations in
this case.
Indeed, it in no way addresses claims
one (Rec1 amino acid sequence fabrication) or two (rec2-1 DNA sequence
falsification) at all. It is topically related to claim three, which
involves what we allege were false statements related to image(s) presented in
a paper produced by the defendants in 1994, and used repeatedly as of
foundational importance to several subsequent grants, including through last
year (2007)." (Sec. Decl. par.
40.)
"Counsel
for the Cornell Defendants have mischaracterized my deposition testimony as
suggesting that I believe that there are no false statements within the Bennett
and Holloman 2001 paper. This is not the case." (Sec. Decl., par. 41.)
"First,
I note that defendant Holloman himself actually backed away from the validity
of the methodology in the Bennett Paper one year after renewal of a $1.7
million grant for project GM42482, i.e.
GM42482-12A2. In that competitively
renewed, and twice amended grant, Holloman claimed, “We have only just recently succeeded in being
able to produce sufficient amounts of both Rec2 and Rad51 …” (p. 14)
[at RPG 00842]. However, in the
next year’s Progress Report, it is stated that, ““Isolation of Rec2 protein has
continued to be a formidable problem
… yields of active protein were low and the
method was not reliable.”" (Sec. Decl., par. 42.)
By
the following year, Holloman effectively declared that the Bennett procedure
had been abandoned:
"Isolation of Rec2 has
continued to be a formidable problem. ... We have continued seeking a better
system for expression of Rec2 and have pursued our finding that soluble Rec2
could be obtained when the gene was expressed in yeast ..." (GM42482-14,
p.2.)
It
is of significant note that, despite having been served with court
interrogatories and requests for production of documents, Holloman failed to produce any data on Rec2 activity from 2002 or
thereafter to support his contention to the NIH in grant application
GM42482-12A2 that he had "sufficient" active Rec2 to even
"begin" the studies for which he was funded.
In other words, there should have been some data produced after the grant
was funded from the sufficient Rec2 portrayed as already being on hand (or
some explanation given for why such data was not produced).
With
respect to concerns about the irreproducibility of the Bennett protocol, I
further noted:
"It
is my suspicion, for many reasons to be further elaborated, but not germane to
the current motions, that the Bennett and Holloman publication of 2001 is
likely part of a continuing fraud.
Its lack of scientific value in terms of reproducibility are strongly
suggested by Defendant Holloman’s own statements in GM42482-13." (Sec. Decl., par. 46.)
I
obtained Bennett's thesis from public sources. The description of his thesis
work suggested to me one possible explanation was that his results may have
been an artifact of contamination by the bacterial (E. coli) enzyme, helicase II. For reasons not explained, Bennett
did not continue precautions to use bacteria that lacked this activity, even
though in his predecessor's, Rubin's, thesis, a significant warning regarding
this issue had been given.
What
follows are three statements Rubin made in his thesis with respect to the
experience he had with a bacterial contaminant activity while trying to study
the activity of purified Rec2:
"A mock preparation was also
set up in parallel to show that the helicase that had been purified was not
present in the strain that had been used to produced Rec2. A culture of BCM465
which carried the pET3b plasmid only (no REC2
insert) was used to produce a soluble protein lysate as described recently. A
major shock came when the fractions from the heparin agarose column
were assayed from DNA-dependent ATPase activity and the activity profile was
exactly the same as with the lysate prepared from the Rec2 overproducing
strain." (Rubin Thesis, p.77;
emphasis added.)
"To sidestep the need to
purify helicase II away from Rec2, we decided to construct an E. coli expression strain with a uvrD::Tn5 mutation
which encodes helicase II. We also took the opportunity to make the strain endA1 which results in lack of
endonuclease I activity. It was desirable to remove endonuclease I from the
background biochemical activities because many of the assays that we had in
mind for Rec2 involved DNA substrates which would be targets for endonuclease I
degradation." (Rubin Thesis, p.79;
emphasis added.)
"The pitfalls of attempting to
assign a biochemical function to a cloned gene are many. In the case of
"Rec2", generic biochemical activities were probed for. These
included: DNA-dependent ATPase, helicase, ATP binding, and ATPgS binding. Failure to do proper controls,
coupled with such generic assays led to the incorrect assignment of a
helicase activity to Rec2. This illustrates an important limitation of this
approach. If one looks for generic activities such as DNA-dependent ATPases in E. coli, they will be found. Only
through controls and attention to meticulous biochemistry can the
assignment of this type of common activity be assigned to an overexpressed
protein of unknown activity." (Rubin
Thesis, p.82; emphasis added.)
Bennett
acknowledged the same risks in his thesis:

Nevertheless,
rather than follow Rubin's approach to use bacteria with as few active
contaminants as possible, Bennett apparently used an Eschericia bacterial strain
which retained the same helicase activity.
Furthermore,
Bennett relied upon the same sorts of generic assays during purification that
Rubin had warned against.
Bennett
did claim to perform assays intended to find such contaminants, but one has to
wonder why, given the risks and Rubin's experience, Bennett did not at least
attempt to use Rubin's helicase-disrupted strains.
However,
the most troubling lack of experimental control was that Bennett did not employ
in his purifications the one which revealed to Rubin that trouble was afoot - an
expression plasmid lacking the REC2 gene. It was use of this negative
control that led Rubin to his "major shock". Clearly it was available
in the laboratory for Bennett's use, and had been shown to be of great value.
The
problems in producing active Rec2 that had supposedly already been in hand when
the NIH first received the grant, continued even after the Bennett procedure
had apparently failed. Holloman stated in progress reports to NIH that he would
try obtain active Rec2 using yeast expression constructs; however, that, too,
did not work:
"... we have not still not
[sic] been able to purify Rec2 past one or two fractionation steps before it
becomes badly degraded. This disappointing result has led us to reconsidering our strategy."
(GM42482-14, p.3.)
Notably,
under such desperate circumstances in which no experiments were reported having
produced expected data for years, Holloman's reconsideration apparently did not
include having Bennett or Kmiec personally produce active Rec2.
As
noted above, Holloman did not reveal during discovery in the legal case any
Rec2 purifications by his laboratory from 2002 or thereafter, or any assessment
as to why Bennett and Kmiec's protocols were so irreproducible in the hands of
others. This situation was reminiscent of that with the "Rec1"
purported strand exchange protein purification, which remained unattainable by
others in Holloman's laboratory for more than a decade (beginning in the
1980's). When those in Kmiec's postdoctoral laboratory could not reproduce his
data, Kmiec was summoned back to that laboratory by its head, Dr. Abraham
Worcel. Under the observation of the Worcel laboratory, Kmiec apparently could
not reproduce his work either.
Therefore,
it appears that the Bennett protocol was quite the opposite of the
"vindication" of the Holloman's 1994 Rec2 work with Kmiec. Rather, it
seems that the experience of Holloman's personnel in not reproducing the
activity claimed by Kmiec was the only thing reproduced with Bennett's method.
Back
to TOC.
IV. New, fourth allegation of false
statements by Holloman to NIH that he had never been able to study soluble Rec2
After
the third year of grant funding, Holloman reported that his lab had found that
soluble Rec2 could be produced in E. coli
by fusion to maltose binding protein (MBP). The soluble Rec2 "thorn"
had been removed, Holloman declared, but no in vitro activity was mentioned as having
been observed.
Indeed,
Holloman failed to provide any evidence of active Rec2 at any time after
funding of this grant in response to discovery in the legal case, either in
2002 when the his grant first claimed "sufficient" Rec2 "to begin" studies, or up to the
point three years later when the original, "fairly manageable" procedure had been abandoned and
purportedly replaced with one using a different Rec2 (MBP) fusion. To my
knowledge, no publication examining in
vitro activity of such a soluble Rec2 has appeared either.
There
is probably a good reason why Holloman made no report of soluble Rec2 having in vitro activity - if that was the case
in subsequent progress reports to the NIH. The latter were not produced by
Holloman during discovery, so at this time adverse inference is taken, i.e., had any evidence to support active Rec2
existed, then Holloman would have had strong motive to produce it to the Court.
one is entitled by adverse inference to assume that such evidence did not
exist.
More
than suspicions about the final results from soluble Rec2 are at issue.
Holloman
made specific claims to the NIH about not
having previously obtained soluble Rec2 for study. In his 2003 grant GM42482-13
Progress Report he stated:
"In previous investigations we
established that recombinant protein could be highly expressed in bacteria, but
could not be obtained in a soluble form without the use of denaturing solvents."
Holloman had also stated in the original grant:
"Unfortunately, the protein
[Rec2] is produced in insoluble form in E. coli",
Examination
of Holloman graduate student Brian Rubin's 1994 thesis, however, indicates that
soluble Rec2 was the primary form of Rec2 that was first examined (p.71):

Rubin
never denatured Rec2 in these experiments, and there is no indication he ever
used "denaturing solvents".
(See also Rubin Thesis, p.116.)
Therefore,
it is alleged that Holloman made additional false claims regarding his having
obtained and studied soluble Rec2 to the NIH in progress reports associated with
his grant GM42482-12A2.
Unfortunately,
due to severe constraints placed by the Court on the time to perform discovery,
and the failure of my attorneys to act in an expeditious manner to adhere to
the limited time available for discovery, no further evidence on either the
third allegation of falsification of protein activity, or the soluble Rec2
issue, was obtained.
Back
to TOC.
I,
Robert P. Bauchwitz, signed this affidavit on __________ at Hershey, PA.
__________________________
Robert P. Bauchwitz
SUBSCRIBED AND SWORN TO BERFORE ME
on __________ at Hershey, PA.
Back
to TOC.
ENDNOTES
(i) More detail on the ORI handling of the case.
1. The ORI's general failure to investigate
i. Plaintiff's attorney Poserina
email to Assistant U.S. Attorneys David Hoffman and Gerald Sullivan, May 26, 2005:
“I am presuming that we are being
granted additional time to clear up the issues presented in the ORI
letter. I am also concerned,
however, that the only review and investigation given in this case was a
scientific paper review of the complaint and disclosure; but there
does not appear to have been any true investigation of the merits of the claims. For instance, there has been no
investigation to determine what, if anything, did occur at the Harvard lab; just
a statement that this information may be old, etc. As far as I can see, there has been no
attempt to interview Dr. Rubin, who may provide the bulk of the information to
corroborate the complaint[1]. There were, however, comments about
whether or not Dr. Rubin said these things or just suggested them; and without
an interview of Dr. Rubin, even telephonically, this is just mere discussion. This
is not an investigation, but just conjecture."
ii. Attorney Poserina email to
Relator of the same date:
“they [DOJ and ORI] are making a
decision without any investigation; with only a paper discussion of the facts. This is far short of the investigation
required under the FCA”.
iii. Attorney Poserina email to
Relator of July 5, 2005:
"Robert: either call me or
let me know when and where I can call you; got a letter today from [ORI’s]
Dahlberg that was not good; basically
they do not feel they could ever get sufficient evidence to proceed."
2. The ORI's apparent misinterpretation of intent standards
7.
With respect to the concern about proving intent, I noted in my first response
to the ORI that even if no further evidence were forthcoming, the standard for
considering intent was more reasonably and broadly specified by the False
Claims Act of 1986, under which this action was being taken. Intent
Standards of the False Claims Act of 1986 (from the first ORI response memo
of 2005; see ORI documents on the CD):
"What Congress said was “It
is intended that persons who ignore ‘red flags’ that the information may not be
accurate or those persons who deliberately choose to remain ignorant … should
be held liable under the Act. This definition, therefore, enables the government
not only to effectively prosecute those persons who have actual knowledge, but
also those who play ‘ostrich’”. (Helmer, JB, False Claims Act: Whistleblower
Litigation, Third Edition, Lexis-Nexis 2002). Therefore, we do not believe that
the “ORI” standard for proof of “intentionally false” is relevant in this
case.
More
specifically, the False Claims Act of 1986 states:
(b) Knowing and Knowingly Defined.
- For purposes of this
section, the terms ''knowing'’ and ''knowingly'' mean that a person, with
respect to information - (1)
has actual knowledge of the information; (2) acts in deliberate ignorance of
the truth or falsity of the information; or (3) acts in reckless disregard of the
truth or falsity of the information, and no proof of specific intent to
defraud is required."
3. The ORI's error of biology
The
specific example ORI provided for their intent concern, affecting only one of
the three allegations of false statements, was based upon an error of biology
on their part. I brought this to their attention in a written response which is
part of the ORI documents (docket document 90, Exhibit 10; also present on the
CD as two pdf files).
Nevertheless,
the ORI persisted in not responding to Department of Justice (DOJ) requests for
clarification as to whether they had indeed made a error of biology (with their
suggestion that protein translation could be expected to begin at the 5’ end of
a eukaryotic RNA. It would not.) (See also my letter to ORI's John Dahlberg,
present in the ORI documents, doc 90, Exhibit 10).
To
present to the Court expert opinions on the issue as to whether ORI may have
been in error, and to further assess the merit of the case based on the
evidence initially presented to the government, reviews by two science experts
who had assessed the information disclosed to the government, as well as the
ORI report, were submitted. One of the reviewers was a member of the
National Academy of Science and a longtime director of a prominent biomedical
research institution, The Carnegie Institution of Washington D.C., while the
other was a professor at the Mayo Clinic. As their complete reports have been
produced during subsequent discovery, a summary of their comments provided to
the Court on January 17, 2006 are reiterated:
i. The two scientists noted the
following concerning the ORI “science” issue:
Reviewer 1: “The [ORI] argument is
embarrassingly
flawed. The conclusion that “regardless of who is correct on the
sequence data, an initiation codon upstream of the published sequence does seem
to exist” [quote from ORI memo 1] is not supported by the available data
and it is both surprising and disturbing that anyone would come to such a
conclusion based on the available data”.
Reviewer 2 “There is no
evidence that [Defendant] H or anyone in H’s lab identified an ATG that
begins an open reading frame that would produce a protein ... there are many
RNAs made by cells that do not make any protein. Without the sequence of the
RNA no conclusion can be made about that RNAs function. Why would one [the ORI] conclude
that this RNA is ‘initiated through a novel and abnormal mechanism’?”
ii. Regarding their overall view
of the case, as requested by the following question: “Do you feel that the
preponderance of the evidence presented indicates that scientific fraud has
occurred in this case?”
Reviewer 1: “Yes. Data, facts, and descriptions thereof appear to have been
manipulated. Results were willfully ignored and presented with the intent of
misleading grant reviewers as well as others in the scientific community.”
Reviewer 2 “Yes. Evidence suggests that on more than one occasion experiments
were never even done. These scientists have been the subject of refutation not
once, not twice, but three times.” [emphasis added]
Therefore, although these science
experts seriously disagreed with ORI regarding the question of biology, they
agreed with ORI that the allegations had merit.
Even
had I not challenged the ORI’s error, in reality what was at issue were lies
made by the Defendants about the actual data they had in their possession, and
their use of such false statements to obtain federal funding. Alternatives
by which desired proteins might have been produced were irrelevant to
consideration of the fraud or underlying scientific
misconduct. Subsequent discovery strongly supported my contention about the
availability of evidence as well as the scientific misconduct involved.
Return to ORI
Statements and Responses





![]()
![]()
![]()
































